Ю
Size: a a a
2020 March 19
@volokhonsky написал свою первую статью для хабра, с чем мы его и поздравляем! Комментарии там 🔥 https://m.habr.com/ru/post/492628/
Ю
Спасибо @Gregory_Demin Григорию Демину, так же участнику нашего чата, от автора статьи в PS
AS
@volokhonsky написал свою первую статью для хабра, с чем мы его и поздравляем! Комментарии там 🔥 https://m.habr.com/ru/post/492628/
👍
AS
Уважаемый человек в ТвиттОре написал, что после того, как в Штатах нормально стали замерять, кол-во заражённых чуть ли больше, чем в Италии вышло.
AS
В Imperial College of London замутили симуляцию, согласно которой мы все скоро умрём. Ещё один уважаемый (кем-то) человек в ТвиттОре написал, что к лету спада ждать не стоит.
Так и живём (пока ещё).
Так и живём (пока ещё).
R
Статью от Imperial college вроде сильно критиковали все, хотя я не вчитывался
IT
@volokhonsky написал свою первую статью для хабра, с чем мы его и поздравляем! Комментарии там 🔥 https://m.habr.com/ru/post/492628/
AS
Статью от Imperial college вроде сильно критиковали все, хотя я не вчитывался
Дальше слова "симуляция" я читать не стал.
АР
В Imperial College of London замутили симуляцию, согласно которой мы все скоро умрём. Ещё один уважаемый (кем-то) человек в ТвиттОре написал, что к лету спада ждать не стоит.
Так и живём (пока ещё).
Так и живём (пока ещё).
АР
А где можно прочесть про эту симуляцию?
AS
В последнем комменте к посту @volokhonsky на хабре кто-то постил. У меня в твиттере мелькнуло и я даже не лайкнул, так что теперь не найдёшь.
БА
Други, кто по Shiny мастер?
Есть DataRangeInput с выбранным диапазоном с 01 марта по 15 марта. Но данные эта зараза фильтрует с 02 марта по 14 марта.
Почему так может быть?
Есть DataRangeInput с выбранным диапазоном с 01 марта по 15 марта. Но данные эта зараза фильтрует с 02 марта по 14 марта.
Почему так может быть?
A
Посмотрите на правила фильтрования >= <=, у вас там может быть > и <
БА
А[
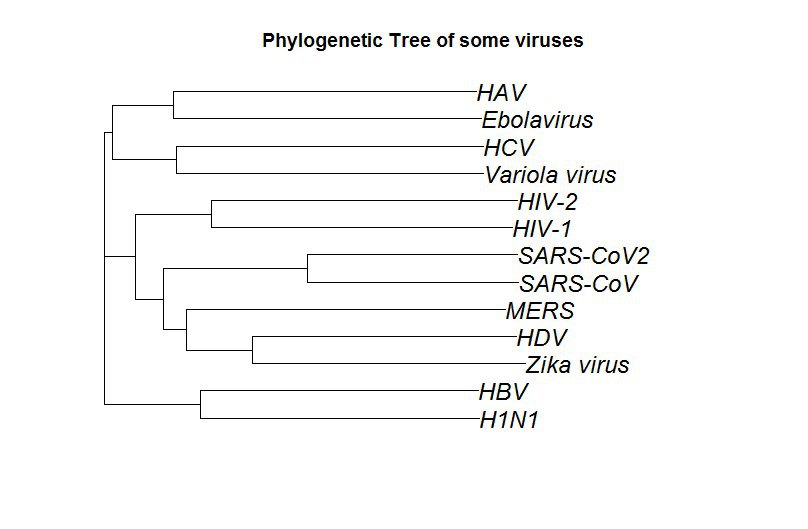
Может кому пригодится, может ошибку найдете. Тут, технический персонал на кафедре поднял вопрос о том, что SARS-CoV2 искусственно выведен в секретных лабораториях, мол там и ВИЧ внутре, и еще много чего, и крч это вообще вирус-убийца, который запрограммирован на уничтожение человечества. К моему ужасу инфа эта была от кого-то с кафедры медбиологии, поэтому захотелось ответить аргументировано. Файло все скачано с базы данных нуклеотидных последовательностей, кодец нагуглен за минуту, считалось долго, поэтому картиночку прилагаю
А[
# install.packages("BiocManager")
# BiocManager::install("msa")
# https://www.ncbi.nlm.nih.gov/nuccore/NC_045512.2?report=fasta # SARS-CoV2
# https://www.ncbi.nlm.nih.gov/nuccore/AF033819.3?report=fasta # HIV-1
library("msa"); library("seqinr"); library(ape)
SARSCoV <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/SARSCoV.fasta")
MERS <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/MERS.fasta")
SARSCoV2 <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/SARSCoV2.fasta")
HIV1 <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/HIV1.fasta")
HIV2 <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/HIV2.fasta")
Variola <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/Variola virus.fasta")
H1N1 <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/H1N1.fasta")
Zika <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/Zika.fasta")
Ebola <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/Ebola.fasta")
HAV <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/HAV.fasta")
HBV <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/HBV.fasta")
HCV <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/HCV.fasta")
HDV <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/HDV.fasta")
da <- msa(inputSeqs = c(SARSCoV, MERS, SARSCoV2, HIV1, HIV2, Variola, H1N1, Zika, Ebola, HAV, HBV, HCV, HDV))
save(da, file = "D:/Work/Projects/SmallSciTheses/2019ncov/da")
da2 <- msaConvert(da, type="seqinr::alignment")
da2$nam <- c("HIV-1", "HIV-2", "SARS-CoV", "SARS-CoV2", "Zika virus", "HDV", "MERS", "Variola virus", "HCV", "Ebolavirus", "HAV", "H1N1", "HBV")
da.dist <- dist.alignment(da2, "s")
dt <- nj(da.dist)
plot(dt, main="Phylogenetic Tree of some viruses", cex = 1.5)
# BiocManager::install("msa")
# https://www.ncbi.nlm.nih.gov/nuccore/NC_045512.2?report=fasta # SARS-CoV2
# https://www.ncbi.nlm.nih.gov/nuccore/AF033819.3?report=fasta # HIV-1
library("msa"); library("seqinr"); library(ape)
SARSCoV <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/SARSCoV.fasta")
MERS <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/MERS.fasta")
SARSCoV2 <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/SARSCoV2.fasta")
HIV1 <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/HIV1.fasta")
HIV2 <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/HIV2.fasta")
Variola <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/Variola virus.fasta")
H1N1 <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/H1N1.fasta")
Zika <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/Zika.fasta")
Ebola <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/Ebola.fasta")
HAV <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/HAV.fasta")
HBV <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/HBV.fasta")
HCV <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/HCV.fasta")
HDV <- readAAStringSet(filepath = "D:/Work/Projects/SmallSciTheses/2019ncov/HDV.fasta")
da <- msa(inputSeqs = c(SARSCoV, MERS, SARSCoV2, HIV1, HIV2, Variola, H1N1, Zika, Ebola, HAV, HBV, HCV, HDV))
save(da, file = "D:/Work/Projects/SmallSciTheses/2019ncov/da")
da2 <- msaConvert(da, type="seqinr::alignment")
da2$nam <- c("HIV-1", "HIV-2", "SARS-CoV", "SARS-CoV2", "Zika virus", "HDV", "MERS", "Variola virus", "HCV", "Ebolavirus", "HAV", "H1N1", "HBV")
da.dist <- dist.alignment(da2, "s")
dt <- nj(da.dist)
plot(dt, main="Phylogenetic Tree of some viruses", cex = 1.5)
2020 March 20